We want to approximate $f: \mathcal{X} \to \mathcal{Y}$, $\mathcal{X} \in \mathbb{R}^p$, $\mathcal{Y}={1,…,L}$
We note:
$x=(x_1,…,x_p)^T \in \mathcal{X}$ a sample
The distance between 2 samples: $d: \mathbb{R}^p \times \mathbb{R}^p \to \mathbb{R}$
$\mathcal{D}_n={(x_i,y_i),i=1,…,n}$ the training set with $n$ samples and labels.
For each new sample $x \in \mathbb{R}^p$, we determine the set of k nearest neighbors among all the train set.
Note: a sample is an observation, that is $x^T=(x_i^{(1)},…,x_i^{(p)})$. We thus compute the distances between vectors of dimension $p$.
The most basic distance metric is the Euclidean norm: $d(u,v)=||u-v||^2=\Sigma_{i=1}^p(u_i-v_i)^2$
The algorithm consists in building a distance matrix with the test sample in columns (power $(t)$) and the train sample in rows (power $(T)$) as shown below:

Note: in this matrix, the training sample is of size $n$ and the test sample is of size $m$.
We then sort the matrix column-wise in ascending order in order to find train data points with smallest distance to the test data point.
For each test data point, we keep the $k$ smallest distances (i.e. the $k$ first rows of the sorted matrix) and we look at their labels. The Python function argsort() allows us to keep indices when sorting. We define the rank of a neighbor as:
\(r_k(x)=i^* \text{ if and only if:}\) \(d(x_{i^*},x)=\underset{1 \le i \le n \\ i \neq r_{1},...,r_{k-1}}{\operatorname{min}}d(x_i,x)\)
The most basic rule is to take the most frequent label among the $k$ neighbors. In the below schema we consider $k=5$ and $k=11$ with three classes ($L=3$) that are drawn respectively in black ($y=1$), grey ($y=2$) and white ($y=3$).
\[\widehat{f}_k(x)=\underset{y \in \mathcal{Y}}{\operatorname{argmax}}(\Sigma_{j=1}^k \unicode{x1D7D9}\{y_{r_j}=y\})\]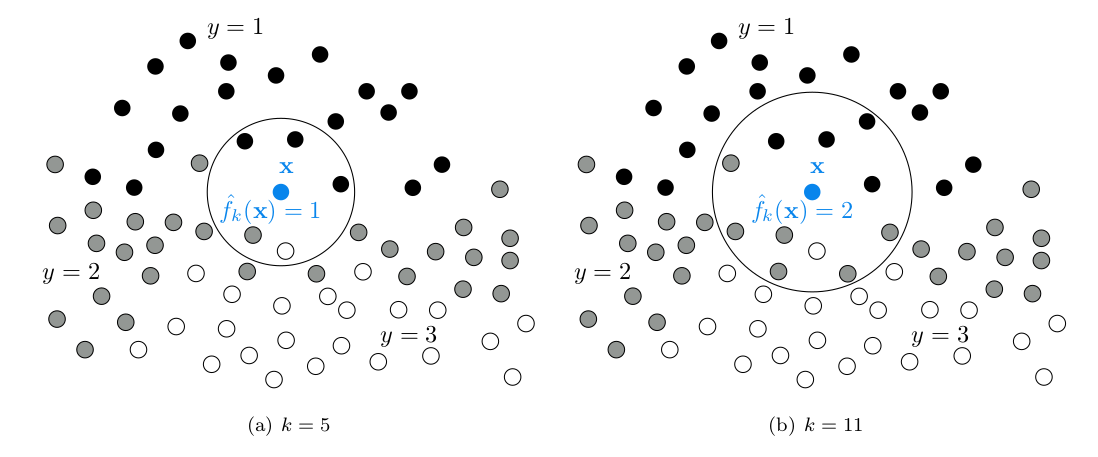
Note: the fit() function of the KNNClassifier is just an affectation of the training data to the object; the distance computation is done in the predict().
The biggest advantages of the KNN algorithm are:
-
It’s a simple and intuitive algorithm that can be easily explained and understood.
-
Its parameter can be easily optimize e.g. using cross validation.
The biggest drawbacks are:
-
The prediction can be computationally expensive because we need to compute all distances between test and train data.
-
It has a high variance: the prediction can be strongly different if the data are slightly different.
Alternative
\[\widehat{f}_k(x)=\underset{y \in \mathcal{Y}}{\operatorname{argmax}}(\Sigma_{j=1}^k \omega_j \unicode{x1D7D9}\{y_{r_j}=y\})\] \[\text{where:}\] \[\omega_j = e^{-d_j^2/h}\]This alternative doesn’t change the knn selection, it only changes the class attribution in the neighborhood. It gives more weights to very small distances. The higher $h$, the higher we favor small distances ($exp$ function becomes steeper).
class KNNClassifier(BaseEstimator, ClassifierMixin):
""" Homemade kNN classifier class """
def __init__(self, n_neighbors=1):
self.n_neighbors = n_neighbors
# the training step only consists in storing training data
def fit(self, X, y):
self.X = X
self.Y = y
return self
def predict(self, X):
n = len(self.X) # size of train set
m = len(X) # size of test set
dist_mat = []
for i in range(n): # we loop on every element of the train set
dist_vect = []
for j in range(m): # we loop on every element of the test set
dist_vect.append(euclidean_distance(self.X[i], X[j]))
dist_mat.append(dist_vect)
# len(dist_vect) = m (nb of features; all the distances for one observation)
dist_mat = np.asarray(dist_mat)
# dist_mat.shape = (n,m); T_test in column, X_train in row
# dist_mat = metrics.pairwise.pairwise_distances(
X, Y=self.X, metric='euclidean', n_jobs=1
)
idx_sort = np.argsort(dist_mat, kind='mergesort', axis=0)
# idx_sort.shape = (n,m); dist_mat sort column-wise
# mergesort is a stable way to handle equal numbers:
# if equal, the order of indices in the output is the same as in the input
idx_sort_knn = idx_sort[:self.n_neighbors,:] # resize with the number of knn
return getBestClassFromCount(idx_sort_knn, self.Y)